Author: Gao Yanxuan
On June 4th, Cell published online the latest research paper by Professor Liu Zheng’s team from The Institute for Advanced Studies of Wuhan University, Taikang Center for Life and Medical Sciences, and Hubei Key Laboratory of Cell Homeostasis, and Professor Zhang Xinghua’s team from the College of Life Sciences, Wuhan University. This paper is titled ‘DNA-based ForceChrono Probes for Deciphering Single-Molecule Force Dynamics in Living Cells’. Dr. Hu Yuru and Dr. Li Hongyun from the Institute for Advanced Studies, Dr. Zhang Chen from the College of Life Sciences, and Feng Jingjing, a doctoral student from the Institute for Advanced Studies, are listed as the paper’s co-first authors. The corresponding authors are Professor Liu Zheng and Dr. Li Hongyun from the Institute for Advanced Studies and Taikang Center for Life and Medical Sciences, and Professor Zhang Xinghua from the College of Life Sciences. Wuhan University is the sole affiliated institution for this publication.

Top row: Zhang Chen, Zhang Xinghua, Liu Zheng, Hu Yuru, Feng Jingjing
Bottom row: Liu Xinping, Chen Wei, Yu Miao
(from left to right)
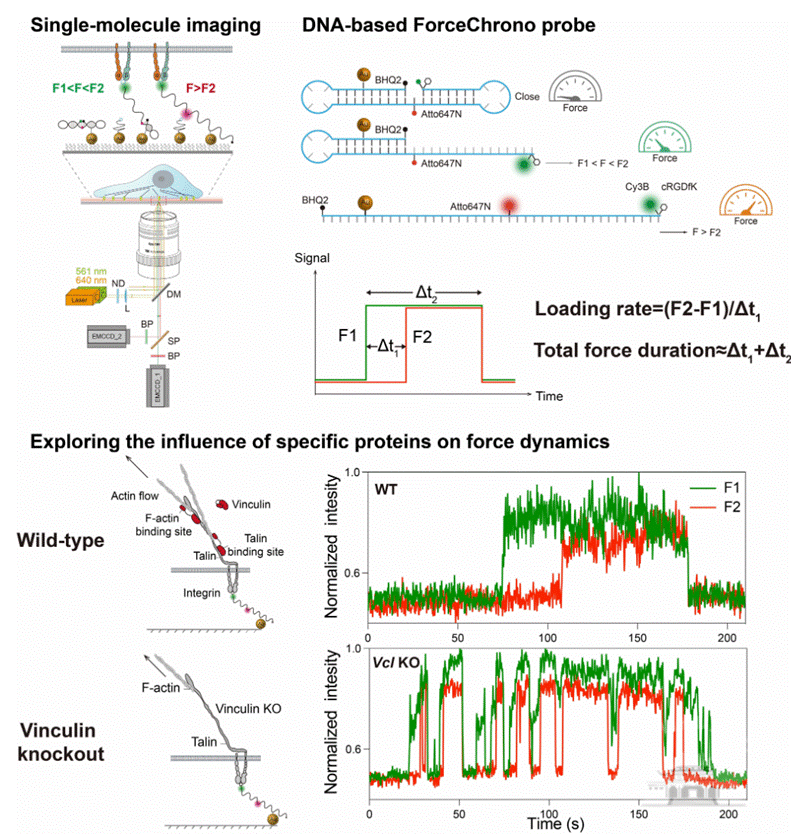
Mechanical force plays a crucial role in cellular signal transduction, distinct from traditional biochemical and electrophysiological signals. The forces transmitted by cell surface receptors are extremely small, ranging from a few to tens of piconewtons (pN), making their dynamic characteristics challenging to measure. Besides the magnitude of the force, its duration and loading rate also profoundly impact cellular signaling and behavior. The duration of the force determines the length of signal transduction, while the loading rate affects protein unfolding and activation. Previous literature and in vitro experiments estimate a broad range of mechanical loading rates, from 0.007 to 10^6pN/s. However, there is a significant lack of understanding regarding the duration of force transmission within living cells. This gap often forces researchers to make rough assumptions about the rate or duration when interpreting phenomena, leading to confusion in understanding cellular mechanotransduction.
To address these challenges, the joint research team of Liu Zheng and Zhang Xinghua developed a novel tool called the ForceChrono Probe. This probe utilizes a pair of DNA hairpin structures, meticulously designed to respond differentially to the pN-level mechanical forces transmitted through membrane proteins. Using single-molecule fluorescence imaging technology, the team achieved a pioneering feat by simultaneously measuring the force transmission duration, mechanical loading rate, and force magnitude on individual integrin molecules within living cells. This breakthrough provides unprecedented insights into the dynamics of force application at the single-molecule level in a cellular context. Moreover, by analyzing how these mechanical parameters are affected by protein mutations, deletions, or pharmacological interventions, the researchers can deduce the functional roles of specific proteins or domains in the complex process of cellular mechanotransduction. This approach allows for a deeper understanding of the intricate roles different proteins play in the mechanical signaling pathways within cells.
Therefore, this work not only provides a novel tool for unveiling the dynamic characteristics of cellular mechanical forces and cellular mechanosensing behavior at the single-molecule level but also suggests that some single-molecule force spectroscopy data need to be reinterpreted considering the physiological loading rates and force durations.
It is reported that this work was supported by the Original Exploration Program and General Program of the National Natural Science Foundation of China (NSFC).
Link to the paper: https://www.cell.com/cell/fulltext/S0092-8674%2824%2900517-8
DOI:https://doi.org/10.1016/j.cell.2024.05.008
Rewritten by Chen Aojie, Edited by Ma Shuqi