Since outbreak of novel coronavirus pneumonia (COVID-19), the confirmed cases have reached 80,000 worldwide on March 1, with the death toll climbing to about 3000. A large number of patients remain on clinical suspicion. WHU joint team has now devolved an original test method, Nanopore Targeted Sequencing test (NTS), which significantly improves the test efficiency. Also, it helps identify SARS-CoV-2 and other 10 categories, 40 kinds of common respiratory viruses simultaneously and monitor mutation at the same time.
Beforehand, qPCR nucleic acid test was the mainstay of COVID-19 diagnosis. However, it shows high false-negative rate and low sensitivity and its positive rate is between 30% and 50%. As a result, numerous highly suspected cases and false recovery cases in latent period are tested false negative. In addition, qPCR nucleic acid test fails to distinguish SARS-CoV-2 from other similar respiratory virus frequently occurring during autumn and winter, thus throwing challenges to triage decision making in the epidemic prevention and control.
Experts on the front line are anxious and eager for a more effective and efficient testing method. Professor Liu Tiangang from WHU School of Pharmaceutical Sciences, Professor Li Yan, Professor Yu Lilei from Renmin Hospital of WHU, Dr. Fu Aisi, the director of Wuhan Dgensee Clinical Laboratory Co., Ltd. formed a joint team along with other experts to develop a new method—Nanopore Targeted Sequencing test (NTS). NTS combines advantages of virus target amplification, long-read nanopore sequencing and real-time data output, which enables SARS-CoV-2 and other 10 categories, 40 kinds of common respiratory viruses to be sensitively and accurately detected simultaneously within four hours for the first time. Its detection limit is 100 times more sensitive than that of the commonly used qPCR. Meanwhile, NTS can also test mutated genome of SARS-CoV-2 and monitor the subsequent virulence and transmission change.
On March 3, the team published their preprint article titled Nanopore target sequencing for accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses on medRxiv.

The research team tests SARS-CoV-2 with NTS at designated hospital in Wuhan
NTS method has gone beyond the sites recommended by Chinese Center for Disease Control and Prevention or American Centers for Disease Control (CDC) in qPCR methods by adopting a wider detection region that includes 9 genes and 12 sites covering nearly 10 kb. Main gene regions of genome are included and important virulence-related genes are fully targeted as well, thus getting a 100 times wider tested genome range than before. Consequently, NST shows much higher sensitivity and accuracy. In contrast, qPCR, only tests 2 to 3 sites covering less than 0.5% genome. Based on a few sites, qPCR test becomes low-efficiency under circumstances where samples are contaminated during sampling, storage or detection phrase. And undetected genes will lead to false negative results. Moreover, the mutated nucleic acid sequence can cause failure test.
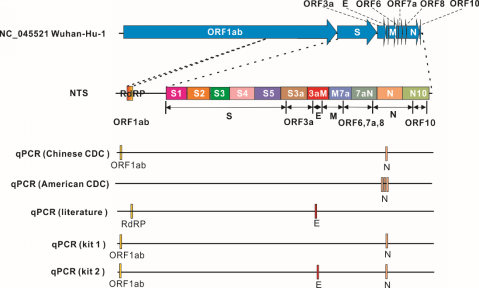
the comparison of NTS and qPCR
According to Professor Liu, qPCR is like a sniper rifle, targeted yet may miss silent genes during samples testing. By contrast, NTS casts a wide net in testing various virus and can also read sequence simultaneously. Therefore, a single test can detect not only SARS-CoV-2, but also respiratory viruses including bocavirus, rhinovirus, human metapneumovirus, respiratory syncytial virus, coronavirus, adenovirus, parainfluenza virus, influenza A virus, influenza B virus and influenza C virus. This provides accurate basis for triage. Furthermore, it can indicate the virulence-related genes mutation during virus spreading, in order to provide instant information for epidemiological analysis.
The team conducts parallel tests on NTS and qPCR. Results show that NTS identifies 34 positive samples out of 45 highly suspected samples, 15 more than qPCR. NTS identifies all the confirmed samples in 16 suspected samples while qPCR 9 only. The clinical samples indicate that NTS contributes to an increase in positive rate at 43.8%. Additionally, NTS takes only 10 minutes to complete the test in high concentration samples while just 4 hours in the low concentration. The whole NTS test procedure will finish in 6 to 10 hours and also the requested nanopore sequencing platform is not highly demanded. The smallest nanopore sequencer (MinION) is mobile and it is suitable for hospitals of all levels.
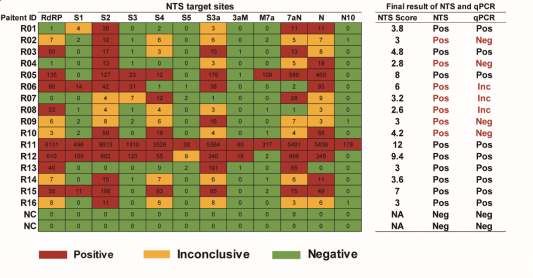
parallel tests of 16 SARS-CoV-2 infected samples
Recently, NTS is of great importance to suspected cases diagnosis as well as epidemic prevention and control thanks to its sensitivity, accuracy, celerity and easy manipulation.
Paper Link: https://www.medrxiv.org/content/10.1101/2020.03.04.20029538v1.full.pdf
Rewritten by: Zhang Yuting
Edited by: Cao Siyi, Shen Yuxi and Hu Siji