On July 21, Advanced Science (IF=15.84) published a full paper on the latest research findings by Professor Liu Tiangang's team from the School of Pharmaceutical Sciences (WHU). Liu’s team developed a cell-free protein synthesis (CFPS) platform which can provide a universal, rapid and efficient method for discovering novel lanthipeptides and guiding their overproduction.
a cell‐free platform for discovering novel lanthipeptides and guiding their overproduction
The paper is entitled “A Cell-Free Platform Based on Nisin Biosynthesis for Discovering Novel Lanthipeptides and Guiding Their Overproduction In Vivo”. Liu Ran, a doctoral student from the School of Pharmaceutical Sciences, is the first author, and Liu Tiangang is the independent corresponding author of the paper. This work was supported by the National Key Research and Development Program of China for Synthetic Biology, the National Natural Science Foundation of China and the Science Foundation for Distinguished Young Scholars of Hubei Province.
The paper investigates the synthesis and post-translational modification of ribosomal peptides (RiPPs), which is a class of peptides synthesized from ribosomes and modified after translation. Lanthipeptide is the most representative compound, which has a wide variety of structures and bioactivities, and a broad range of sources such as nisin. Why doesn’t sauerkraut rot? How can ham sausages be kept at room temperature for months? It's all thanks to nisin’s super bactericidal effect. Nisin can be broken down into amino acids by enzymes secreted by the human body, and has been used for decades as a safe food preservative approved by the WHO and FDA.
Based on the large amount of genomic information, the lanthipeptide compounds that have been discovered and identified only account for a small part of nature, and there are a large number of unknown lanthipeptide compounds to be discovered and elucidated. However, many microorganisms containing the relevant genes cannot be cultured or cannot produce lanthipeptides under laboratory conditions. Although researchers have developed heterologous expression systems such as Escherichia coli and lactic acid bacteria, this method is easy to form inclusion bodies on the one hand, and on the other hand, because most lanthipeptides are bacteriostatic, the expression of lanthipeptides in heterologous host microorganisms becomes a contradiction. The lanthipeptide is a good spear but it pricks the shield (pricks host microbe). In addition, due to the important industrial and medical value of these compounds, there is an urgent need to increase the yield of them. Although the enzymatic mechanism of their biosynthesis is well established, the rate-limiting step of the whole system has not been determined, so it cannot be accurately modified.
Based on the scientific issues above, Liu Tiangang’s group developed a cell-free protein synthesis (CFPS) platform targeting nisin. Although the gene clusters of lanthipeptides are complex, most of them are immune proteins, transport proteins and regulatory proteins that are synthesized by the host to resist its own antibacterial activity, and only a few genes are responsible for the synthesis of lanthipeptides. Therefore, it is simple and efficient to solve the spear-and-shield problem faced in lanthipeptides research.
In this work, the efficient CFPS synthesis platform for nisin was first set up, then the rate-limiting step of its biosynthesis was locked by systematic titration on the CFPS platform, and the metabolic transformation of industrial strains was precisely guided. This step of transformation increased the production of nisin by 60%. This principle was used to guide the metabolic transformation of the heterologous host, which also increased the yield by nearly 90%. Secondly, the platform was used to mine all the potential lanthipeptides similar to nisin skeleton in the NCBI database within one day. In this way, four new lanthipeptides with antibacterial activity (including antibacterial activity against drug-resistant bacteria MRSA) were identified, among which one was superior to nisin in antibacterial activity against clinical pathogenic bacteria enterococcus faecalis. Thirdly, a library of nisin mutants was constructed, and high-throughput screening was carried out through the built CFPS platform. Finally, a lanthipeptide M5 with 5 mutation sites was selected, which endowed this compound with anti-gram-positive bacteria activity. This result can provide a universal, rapid and efficient method for high-yielding lanthipeptides and mining novel lanthipeptides. Also, the formed CFPS platform can also be applied to the research of other RiPPs.
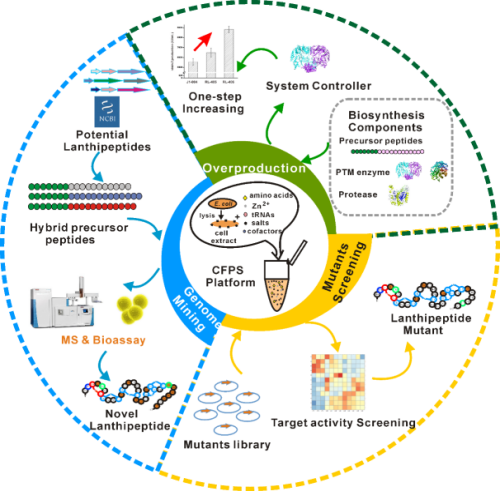
a cell-free protein synthesis platform targeting nisin
links to the paper: https://onlinelibrary.wiley.com/doi/full/10.1002/advs.202001616
links to Liu Tiangang’s research team: http://liugroup.whu.edu.cn/
Written by: Yao Yuan
Rewritten by: Qin Zichang
Edited by: Wu Buer