A research team led by Miao Xiaoping and Tian Jianbo from Wuhan University (WHU) recently published a study in Cancer Discovery. The team utilized single-cell sequencing technology to create a panoramic view of the dynamic evolution of colorectal cancer (CRC) and identify key molecular markers.
The WHU team's paper, Single-cell eQTL mapping reveals cell subtype-specific genetic control and mechanism in malignant transformation of colorectal cancer, in Cancer Discovery.
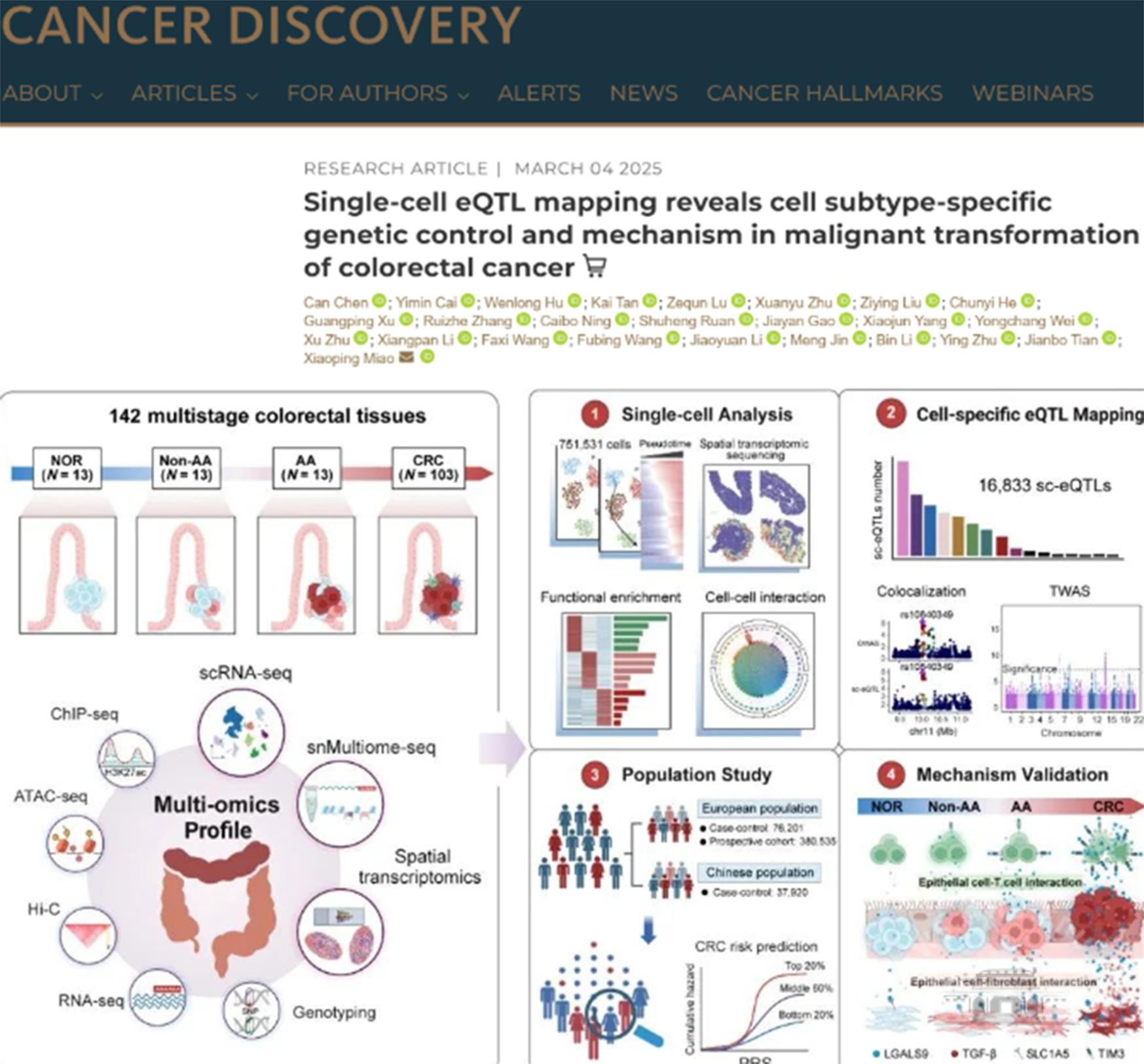
CRC develops from normal tissue to precancerous lesions (Non-AA and AA) before advancing to cancer, offering a crucial window for early detection and intervention. However, cellular heterogeneity and early molecular events in tumor evolution remain unclear.
The WHU team analyzed 142 samples from 102 patients using single-cell RNA sequencing (scRNA-seq), single-nucleus multi-omics sequencing (snMultiome-seq), and spatial transcriptomics (ST). They identified dynamic changes in 48 cell subtypes and found that cell interaction networks become increasingly disordered as the disease progresses, involving inflammatory immune regulation and tumor proliferation pathways.
For the first time, researchers constructed a single-cell expression quantitative trait loci (sc-eQTL) map, identifying 16,833 associations. They discovered that most loci are cell-type specific and enriched in CRC susceptibility regions. A polygenic risk score (PRS) model based on sc-eQTL significantly improved CRC risk prediction, outperforming traditional models. Validation in the Chinese population showed that individuals with high genetic risk were 1.34 times more likely to develop cancer.
The study also revealed that the rs4794979 locus enhances LGALS9 expression, activating cancer-associated fibroblasts (CAFs) and accelerating cancer progression. These findings provide a vital foundation for precision screening and early intervention in CRC.