On March 13, Nature Communications published online a significant achievement entitled “Cell lineage-specific transcriptome analysis for interpreting cell fate specification of proembryos” by Prof. Sun Mengxiang's team from College of Life Sciences, Wuhan University, which reveals the molecular mechanism that determines the fate of cells in early embryogenesis.
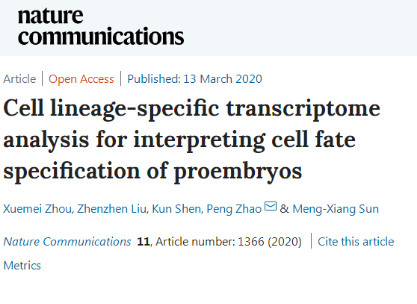
The plant zygotes usually undergo an asymmetric division to form two daughter cells, namely a smaller apical cell and a larger basal cell. The smaller apical cells will divide and develop into the main part of the embryo, while the larger basal cell will undergo few cell divisions to form the embryonic stalk. Therefore, the two daughter cells formed by zygote division are significantly distinct in developmental pathway and developmental fate. However, the molecular mechanisms that regulate the differentiation of parietal and basal cells remain poorly understood.
The researchers created a unique technique for the separation of apical and basal cell lines in early embryo of arabidopsis thaliana, isolated and collected apical and basal cell lines in different developmental stages, and constructed the first international specific transcriptome database of apical and basal cell lines in arabidopsis thaliana in combination with the single-cell sequencing technology, and comprehensively compared and analyzed the differentiation process of apical and basal cell lines in early embryogenesis at the genomic level. Study finds that selective transcription degradation and specific de novo transcription synthesis are involved in cell line specific regulatory pathways to regulate apical and basal cell fate specialization. Since the embryo stage, the apical cell line has specifically activated some molecular regulatory pathways related to embryogenesis, while the basal cell rapidly reconstruct its transcriptome, specializing to the fate of the embryonic stalk, and form a unique gene expression profile. In addition, study finds that many factors such as long non-coding RNA and differential splicing may be involved in determining the fate of apical and basal cells.
This is another important finding by Sun Mengxiang's team in the field of plant embryo development after "transcriptome before and after fertilization", which was published on Developmental Cell in 2019. Post doctor student Zhou Xuemei is the first author, and Associate Professor Zhao Peng is the responsible author. Frasergen participated in the sequencing analysis of transcriptome. PhD student Liu Zhenzhen and Shen Kun also took participation in the research, which is funded by the National Natural Science Foundation of China.
It is worth mentioning that the team recently published a paper on Autophagy entitled Autophagy-mediated compartmental cytoplasmic deletion is essential for pollen germination and male fertility, expounding that autophagy-mediated selective cytoplasmic clearance is a key step in pollen initiation. Zhao Peng is the first author, and Sun Mengxiang is the responsible author. Postdoctoral student Zhou Xuemei, PhD Zhao Linlin and Prof. Alice Y. Cheung from University of Massachusetts, Amherst, USA, also participated in the work funded by the National Natural Science Foundation of China.
The pollen tube produced by pollen germination is one of the key processes in plant sexual reproduction. Previous studies have found that mature pollen grains need two main processes, namely adhesion and hydration, to germinate into pollen tubes. When the dormant pollen grains fell on the stigma, they will absorb water to activate and reorganize their cytoplasm to support the smooth pollen germination. However, the processes activated by pollen hydration have been poorly reported.
The researchers find that autophagy-mediated selective cytoplasmic clearance is another important step in pollen initiation. They used a variety of experimental techniques to confirm that during the pollen germination process, the autophagic activity is significantly enhanced at the pollen germination pore. When autophagy activity is inhibited, a layer of undegraded cytoplasm will accumulate at the pollen germination pore, resulting in the failure of pollen tube to grow out of the germination pore, revealing that this new regional autophagy mode is an essential step for pollen germination. Some cytoplasm in pollen cells must be cleared by this new autophagy to initiate germination.
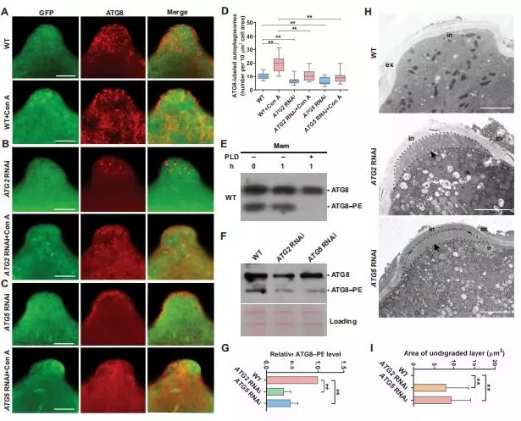
In addition, the comparative analysis of high-throughput omics suggests that autophagy may regulate the pollen germination process through the downstream lipid and mitochondrial related pathways. These findings reveal a critical role for autophagy in initiating pollen germination. This is the first study to report that autophagy plays a role in the development of male gametophytes, revealing a new important link in pollen germination.
Link to the paper: https://www.nature.com/articles/s41467-020-15189-w
Doi: 10.1038/s41467-020-15189-w
Rewritten by: Liu Xinfan
Edited by: Wu Buer, Shen Yuxi and Hu Sijia